IV
09/2020 - 01/2021
COMPUTATIONAL FOREST
The Computational Forest, reforestaion of
a former quarry in Arboç, Spain
Personal Research
09/20 - 01/21

The quarry, located in Arboç within the Garraf National Park, has gained
notoriety for its vast open-air lime extraction quarries. The prospect of
reclaiming such a sizable tract of land presents a remarkable yet intricate
challenge. To make this endeavour possible, it necessitated an indepth
understanding of the intricacies of the natural world.
This undertaking demanded more than a mere reclamation effort; it required a
profound comprehension of ecological dynamics and the delicate balance of ecosystems.
Only through this comprehensive understanding was the proposal to rehabilitate
this extensive landscape made feasible. The restoration of this land holds promise
not only for environmental recovery but also as a testament to the symbiotic
relationship between nature’s resilience and human stewardship. Fungal network
50 years after reforestation intervention
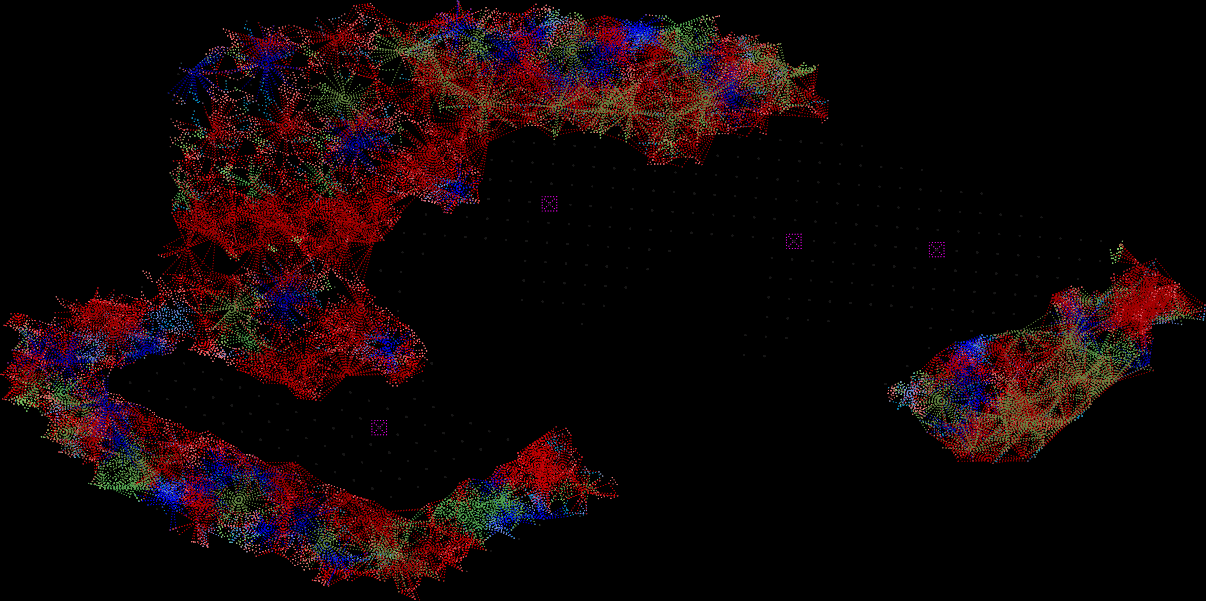
Existing Fungal Network
200 ha of an abandoned quarry
surronded by forest.
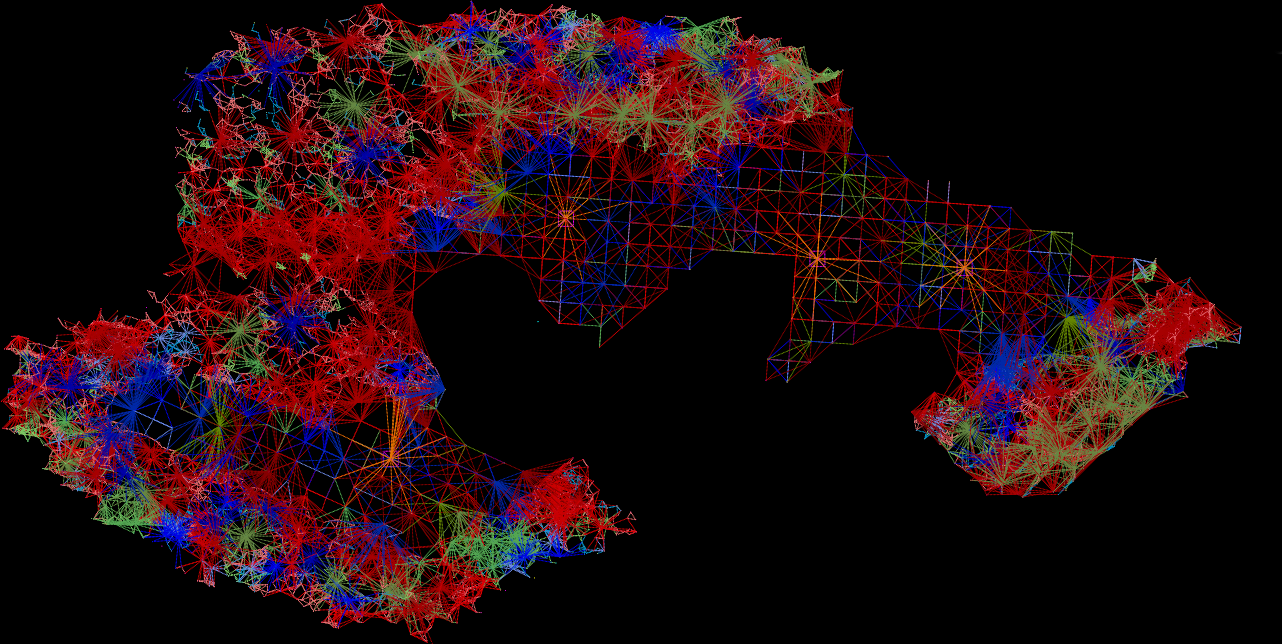
Fungal network 10 years after
reforestation intervention
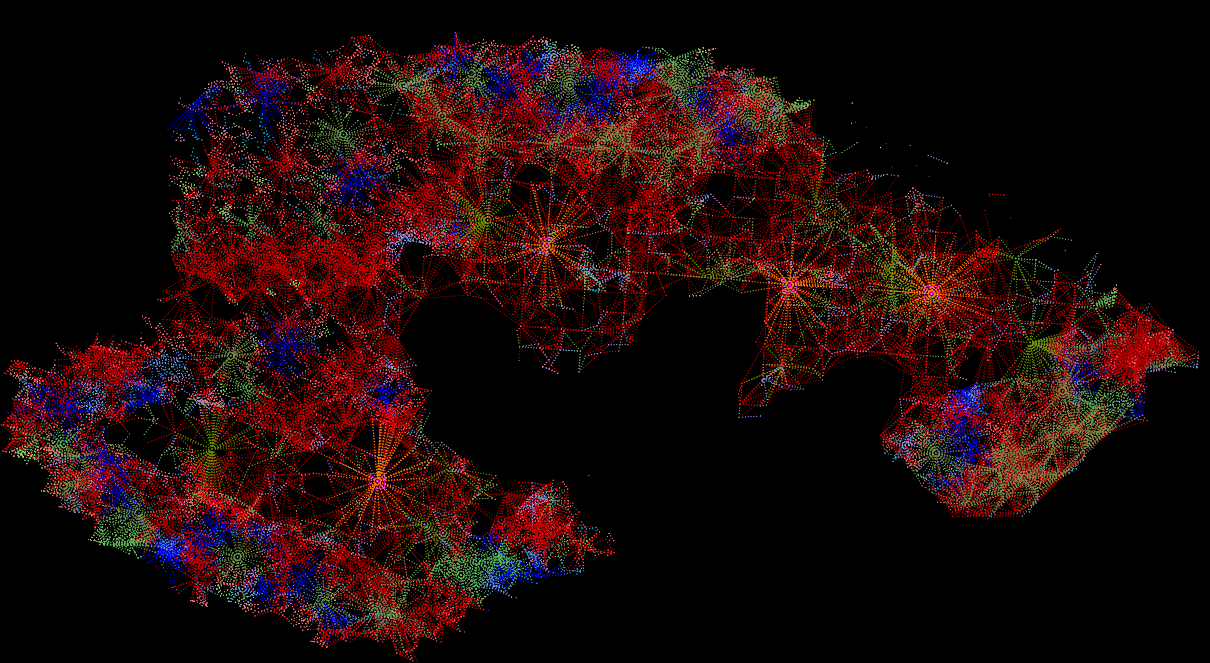
Fungal network 50 years after
reforestation intervention
The computational aspect of the project was done via a custom AUTOCAD LISP. This code explores the creation of connections between trees, defining connection ratios based on tree attributes like species, size, and distance. Through iterations, it compares trees, calculating distances and determining probabilities based on similarities in attributes. The code assesses conditions, including distance thresholds and random probabilities, to decide whether to draw connections between trees. Ultimately, it visually represents these connections as lines in the AutoCAD environment, evaluating and establishing relationships based on predefined criteria.
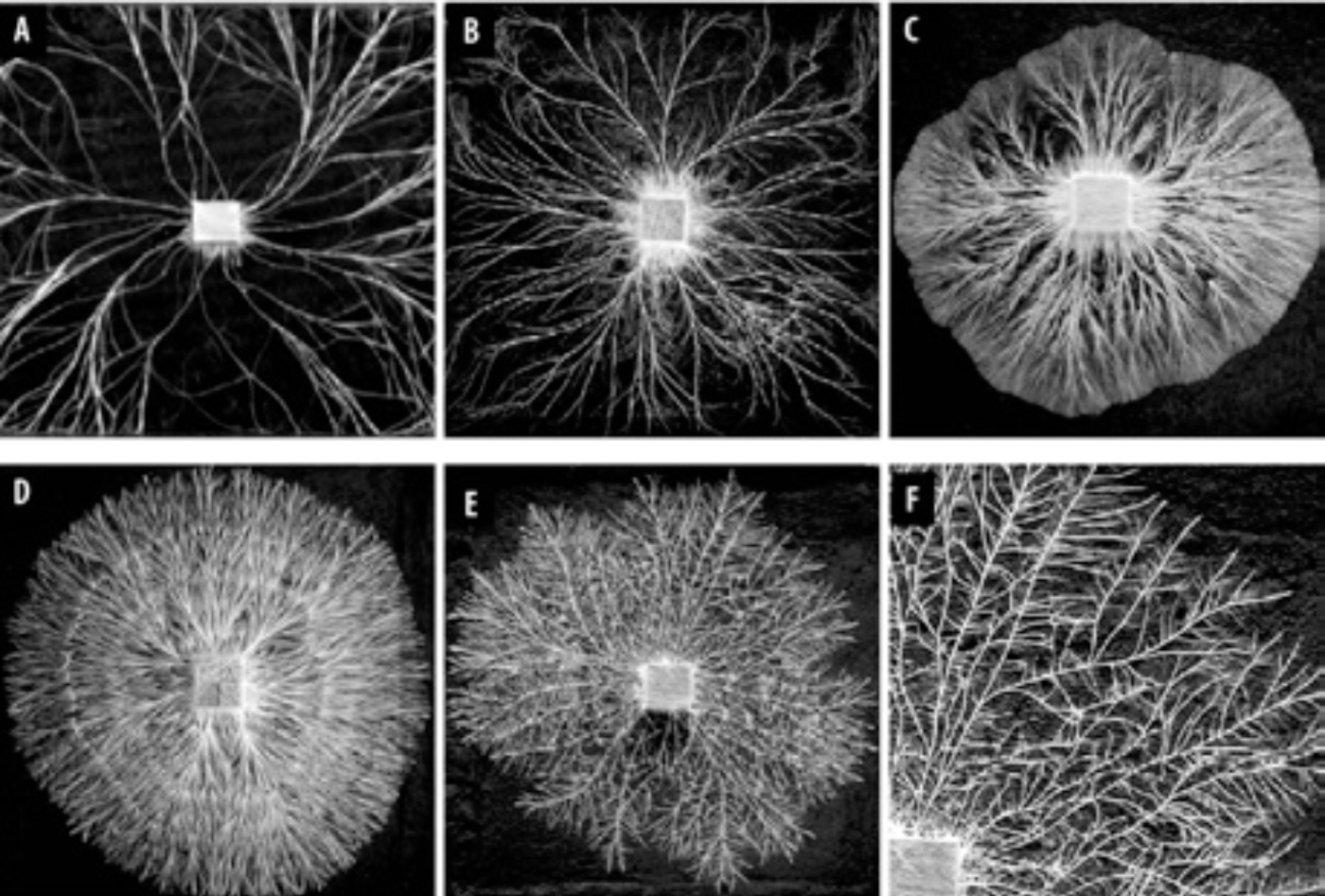
Mycellium growth lab tests
AutoLisp code for the network calculation
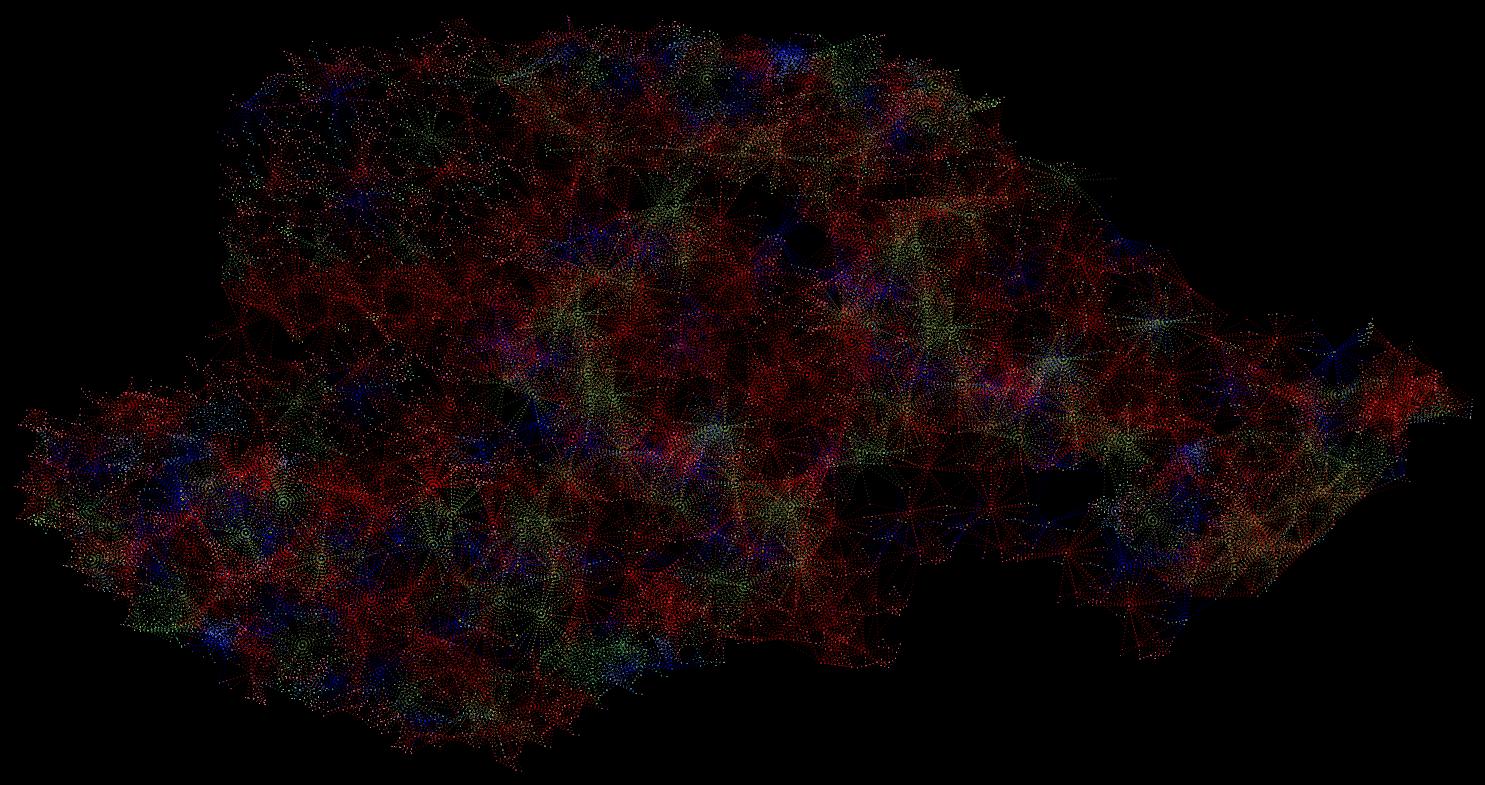
This drawing reveals the full recovery of the fungal network after healing from the scar of the quarry. Distinguished by various colors representing different tree species, the visualization vividly illustrates the intricate connections woven among them.